17. August 2015
Microbial transglutaminase for site-specific protein conjugation
by Roger Schibli (ETH Zurich, Switzerland) and Philipp Spycher (PSI Villigen, Switzerland)
Many techniques have recently been developed to attach functional molecules to proteins either non-specifically or site-specifically. Particularly in the field of antibody-drug conjugates (ADCs) for therapeutic applications site-specific attachment of toxic compounds is a hot topic.
Using site-specific conjugation the ratio protein to attached molecule can be precisely controlled, resulting in conjugates with well-defined stoichiometry. This, for example, avoids conjugation to critical protein interaction sites and thus sustains the binding affinity of the protein. For ADCs, this results in increased tumor uptake compared to heterogeneously modified ADCs.1 As a site-specific technology, enzymatic conjugation has shown great interest since those conjugation reactions are typically fast and can be done under physiological conditions. Among the available enzymes, the microbial transglutaminase (MTG) from the species Streptomyces mobaraensis has recently been recognized as an interesting alternative to conventional chemical conjugation of chemical entities to proteins including antibodies. The MTG catalyzes under physiological conditions a transamidation reaction between a ‘reactive’ glutamine of a protein and a ‘reactive’ lysine residue whereas the latter can also be a simple, low molecular weight primary amine such as a 5-aminopentyl group.2 For an endogenous glutamine on a protein to be recognized as an MTG-substrate two criteria seem important:
1) hydrophobic amino acids around the glutamine seem to foster conjugation3;
2) the glutamine must be situated on a loop with local chain flexibility enhancing reactivity toward MTG.4
From our experience as well as from others it seems, however, that only few glutamines typically fulfill these requirements5 e.g. in antibodies, thus, making the MTG an attractive tool for site-specific and stoichiometric protein modifications.
MTG mechanism10
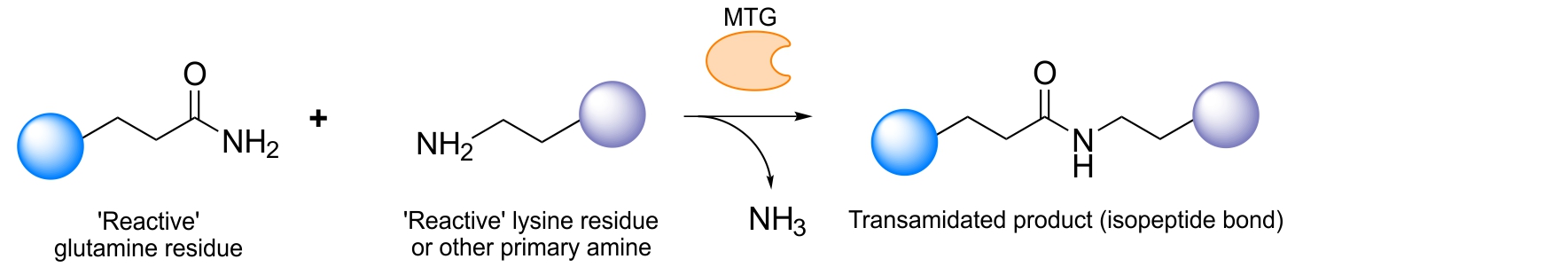
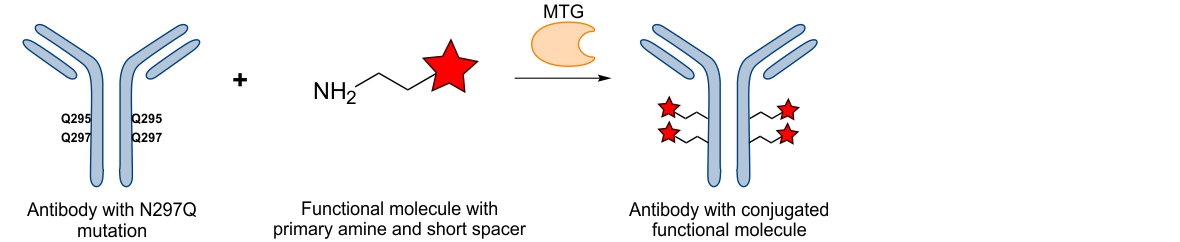
The Schibli group identified a single reactive glutamine on the heavy chain of different IgGs, which is recognized by MTG: Q295. Located on a loop which upon enzymatic deglycosylation of the asparagine 297 (N297) becomes flexible can easily and very efficiently be conjugated with various substrates.6 This property allows for direct modification of the antibody in a straightforward manner. By using this approach, exactly two chemical entities can be conjugated to the antibody backbone. We furthermore discovered that by ablating the asparagine 297 through genetic mutation into a glutamine (N297Q) two more substrates could be attached generating an antibody with four covalently and homogeneously attached substrates. Incorporation of four highly toxic drugs is, for example, particularly attractive for the generation of antibody-drug-conjugates where the drug load plays a decisive role in effective tumor killing.
Antibody conjugation using Q295 and mutation N297Q10
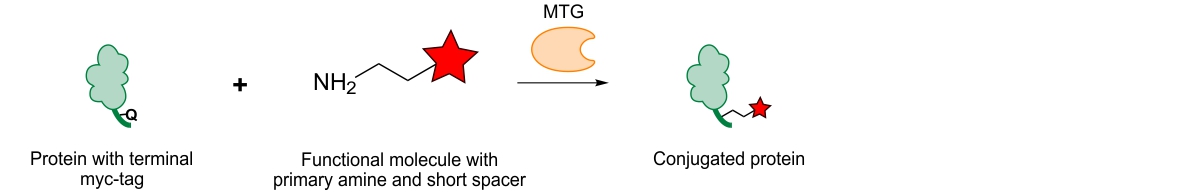
During our studies we have also observed that the genetically introduced myc-tag, consisting of only 10 amino acids with one being a glutamine, was efficiently recognized by the MTG. Upon sequence analysis we could determine that the reaction was indeed catalyzed by the glutamine within the myc-tag.7 Since the myc-tag is often used as a protein identification tool after recombinant protein expression, MTG-mediated bioconjugation can thus be used in a straightforward manner to generate homogenous conjugates with one functional moiety attached at the terminal myc-tag like biotin-cadaverine, a fluorescent dye or a radiochelator. Such a biotinylation can for example be used to immobilize proteins on a solid surface, therefore providing a controlled orientation. In case of immobilized e.g. antibody fragments a controlled orientation will likely enhance interactions with their antigens compared to a random orientation where some of the interactions sites will not be accessible due to an unfavorable orientation. This can also be applied to the uncontrolled conjugation of molecules to proteins since these could as well be attached to important protein-interaction sites leading to decreased binding and lower affinity towards their designated binding partners. In contrast terminal attachment will likely sustain high affinity-binding as such deleterious effects will be avoided. Additionally, attaching several moieties in a non-controlled manner often changes the solubility behavior of the conjugated protein particularly when the attached moieties are bulky and hydrophobic. Altered solubility could lead to unfavorable protein-conjugate aggregation and eventually, precipitation with loss of valuable material. With MTG a controlled amount of the hydrophobic molecule is attached generating conjugates which are well solubilized yet.
Genetic tags like the myc-tag can not only be placed at protein termini but also directly within the protein’s backbone.
Conjugation to proteins using myc-tag10
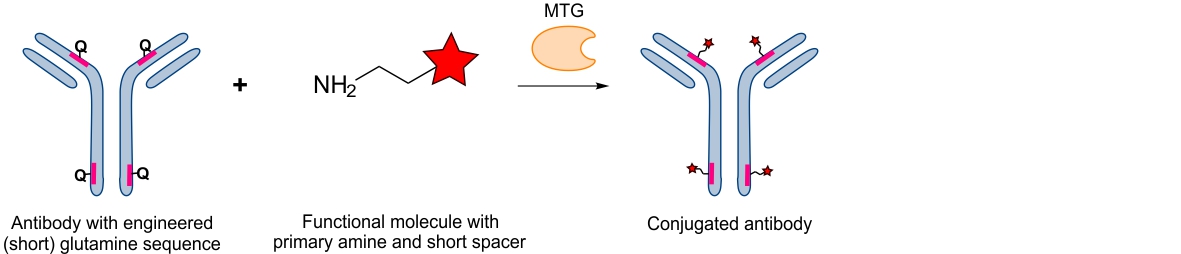
This, for example, has been achieved for antibodies by Pfizer, where the antibody’s amino acid sequence was genetically engineered such that at different positions, probes could be incorporated using MTG and a short four amino acid motif.8 This approach virtually allows conjugating any probe at any position as long as it is accessible enough and the protein structure will not be compromised upon sequence introduction. This approach thus requires careful structure analysis to evaluate and determine the least interfering locations.
Engineered backbone conjugation10
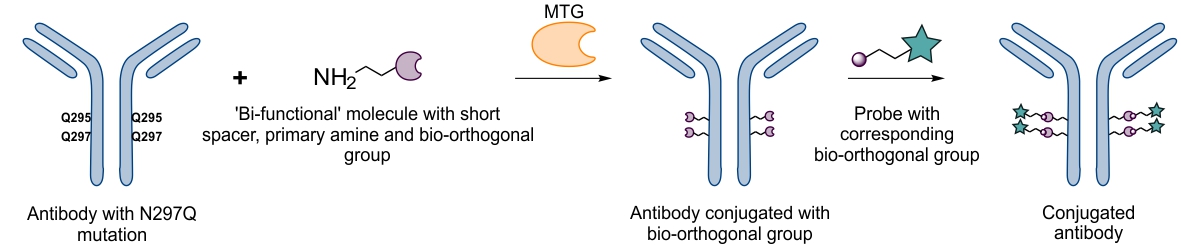
MTG can also be used for chemo-enzymatic functionalization of proteins in case the enzyme is not tolerant to a substrate resulting in a low conjugation yield. Thereby a small ‘bi-functional’ spacer is first conjugated to the protein consisting of a primary amine (recognized by MTG) and a reactive group suitable for subsequent bio-orthogonal coupling of functional molecules. In a second step, and after purification, the functional molecule can easily be conjugated to the already modified protein at low molecular excess. Using this approach, we could conjugate large toxins to antibodies efficiently and at low molecular excess achieving a well-defined stochoimetry.9
Chemo-enzymatic modification10
Taken together, MTG-mediated bioconjugation is a very attractive tool for the efficient generation of homogeneously modified proteins with virtually every substrate as long as it contains a primary amine. However, since MTG is somewhat promiscuous towards the glutamine and lysine residues with no defined substrate preference, it would be desirable to develop MTGs with altered substrate specificities such that different molecules could be conjugated which will be the task for future research.
References:
The authors:


Many techniques have recently been developed to attach functional molecules to proteins either non-specifically or site-specifically. Particularly in the field of antibody-drug conjugates (ADCs) for therapeutic applications site-specific attachment of toxic compounds is a hot topic.
Using site-specific conjugation the ratio protein to attached molecule can be precisely controlled, resulting in conjugates with well-defined stoichiometry. This, for example, avoids conjugation to critical protein interaction sites and thus sustains the binding affinity of the protein. For ADCs, this results in increased tumor uptake compared to heterogeneously modified ADCs.1 As a site-specific technology, enzymatic conjugation has shown great interest since those conjugation reactions are typically fast and can be done under physiological conditions. Among the available enzymes, the microbial transglutaminase (MTG) from the species Streptomyces mobaraensis has recently been recognized as an interesting alternative to conventional chemical conjugation of chemical entities to proteins including antibodies. The MTG catalyzes under physiological conditions a transamidation reaction between a ‘reactive’ glutamine of a protein and a ‘reactive’ lysine residue whereas the latter can also be a simple, low molecular weight primary amine such as a 5-aminopentyl group.2 For an endogenous glutamine on a protein to be recognized as an MTG-substrate two criteria seem important:
1) hydrophobic amino acids around the glutamine seem to foster conjugation3;
2) the glutamine must be situated on a loop with local chain flexibility enhancing reactivity toward MTG.4
From our experience as well as from others it seems, however, that only few glutamines typically fulfill these requirements5 e.g. in antibodies, thus, making the MTG an attractive tool for site-specific and stoichiometric protein modifications.
MTG mechanism10

The Schibli group identified a single reactive glutamine on the heavy chain of different IgGs, which is recognized by MTG: Q295. Located on a loop which upon enzymatic deglycosylation of the asparagine 297 (N297) becomes flexible can easily and very efficiently be conjugated with various substrates.6 This property allows for direct modification of the antibody in a straightforward manner. By using this approach, exactly two chemical entities can be conjugated to the antibody backbone. We furthermore discovered that by ablating the asparagine 297 through genetic mutation into a glutamine (N297Q) two more substrates could be attached generating an antibody with four covalently and homogeneously attached substrates. Incorporation of four highly toxic drugs is, for example, particularly attractive for the generation of antibody-drug-conjugates where the drug load plays a decisive role in effective tumor killing.
Antibody conjugation using Q295 and mutation N297Q10

During our studies we have also observed that the genetically introduced myc-tag, consisting of only 10 amino acids with one being a glutamine, was efficiently recognized by the MTG. Upon sequence analysis we could determine that the reaction was indeed catalyzed by the glutamine within the myc-tag.7 Since the myc-tag is often used as a protein identification tool after recombinant protein expression, MTG-mediated bioconjugation can thus be used in a straightforward manner to generate homogenous conjugates with one functional moiety attached at the terminal myc-tag like biotin-cadaverine, a fluorescent dye or a radiochelator. Such a biotinylation can for example be used to immobilize proteins on a solid surface, therefore providing a controlled orientation. In case of immobilized e.g. antibody fragments a controlled orientation will likely enhance interactions with their antigens compared to a random orientation where some of the interactions sites will not be accessible due to an unfavorable orientation. This can also be applied to the uncontrolled conjugation of molecules to proteins since these could as well be attached to important protein-interaction sites leading to decreased binding and lower affinity towards their designated binding partners. In contrast terminal attachment will likely sustain high affinity-binding as such deleterious effects will be avoided. Additionally, attaching several moieties in a non-controlled manner often changes the solubility behavior of the conjugated protein particularly when the attached moieties are bulky and hydrophobic. Altered solubility could lead to unfavorable protein-conjugate aggregation and eventually, precipitation with loss of valuable material. With MTG a controlled amount of the hydrophobic molecule is attached generating conjugates which are well solubilized yet.
Genetic tags like the myc-tag can not only be placed at protein termini but also directly within the protein’s backbone.
Conjugation to proteins using myc-tag10

This, for example, has been achieved for antibodies by Pfizer, where the antibody’s amino acid sequence was genetically engineered such that at different positions, probes could be incorporated using MTG and a short four amino acid motif.8 This approach virtually allows conjugating any probe at any position as long as it is accessible enough and the protein structure will not be compromised upon sequence introduction. This approach thus requires careful structure analysis to evaluate and determine the least interfering locations.
Engineered backbone conjugation10

MTG can also be used for chemo-enzymatic functionalization of proteins in case the enzyme is not tolerant to a substrate resulting in a low conjugation yield. Thereby a small ‘bi-functional’ spacer is first conjugated to the protein consisting of a primary amine (recognized by MTG) and a reactive group suitable for subsequent bio-orthogonal coupling of functional molecules. In a second step, and after purification, the functional molecule can easily be conjugated to the already modified protein at low molecular excess. Using this approach, we could conjugate large toxins to antibodies efficiently and at low molecular excess achieving a well-defined stochoimetry.9
Chemo-enzymatic modification10

Taken together, MTG-mediated bioconjugation is a very attractive tool for the efficient generation of homogeneously modified proteins with virtually every substrate as long as it contains a primary amine. However, since MTG is somewhat promiscuous towards the glutamine and lysine residues with no defined substrate preference, it would be desirable to develop MTGs with altered substrate specificities such that different molecules could be conjugated which will be the task for future research.
References:
- F. Lhospice, D. Bregeon, C. Belmant, P. Dennler, A. Chiotellis, E. Fischer, L. Gauthier, A. Boedec, H. Rispaud, S. Savard-Chambard, A. Represa, N. Schneider, C. Paturel, M. Sapet, C. Delcambre, S. Ingoure, N. Viaud, C. Bonnafous, R. Schibli and F. Romagne, Mol Pharmaceut, 2015, 12, 1863-1871.
- P. Strop, Bioconjugate Chemistry, 2014, 25, 855-862.
- Y. Sugimura, K. Yokoyama, N. Nio, M. Maki and K. Hitomi, Archives of Biochemistry and Biophysics, 2008, 477, 379-383.
- B. Spolaore, S. Raboni, A. R. Molina, A. Satwekar, N. Damiano and A. Fontana, Biochemistry, 2012, 51, 8679-8689.
- N. Kamiya, T. Takazawa, T. Tanaka, H. Ueda and T. Nagamune, Enzyme Microb Tech, 2003, 33, 492-496.
- S. Jeger, K. Zimmermann, A. Blanc, J. Grunberg, M. Honer, P. Hunziker, H. Struthers and R. Schibli, Angewandte Chemie-International Edition, 2010, 49, 9995-9997.
- P. Dennler, L. K. Bailey, P. R. Spycher, R. Schibli and E. Fischer, Chembiochem, 2015, 16, 861-867.
- P. Strop, S. H. Liu, M. Dorywalska, K. Delaria, R. G. Dushin, T. T. Tran, W. H. Ho, S. Farias, M. G. Casas, Y. Abdiche, D. H. Zhou, R. Chandrasekaran, C. Samain, C. Loo, A. Rossi, M. Rickert, S. Krimm, T. Wong, S. M. Chin, J. Yu, J. Dilley, J. Chaparro-Riggers, G. F. Filzen, C. J. O'Donnell, F. Wang, J. S. Myers, J. Pons, D. L. Shelton and A. Rajpal, Chem Biol, 2013, 20, 161-167.
- P. Dennler, A. Chiotellis, E. Fischer, D. Bregeon, C. Belmant, L. Gauthier, F. Lhospice, F. Romagne and R. Schibli, Bioconjugate Chemistry, 2014, 25, 569-578.
- Figures with courtesy of Philipp Spycher, 2015
The authors:

Prof. Roger Schibli is an Associate Professor in the Department of Chemistry and Applied Biosciences (ETH Zurich) since 2010, and Head of the Center for Radiopharmaceutical Science (CRS), a joint endeavor between the ETH Zurich, the Paul Scherrer Institute and the University Hospital Zurich. Prof. Schibli's research interests cover targeted tumor diagnosis and therapy using radiolabeled compounds. Apart from the chemical and enzymatic modification and radioactive labeling of molecules, his group emphasizes on the biological and pharmacological characterization and optimization of the radioactive compounds including non-invasive PET and SPET imaging.
More information about Prof. Schibli and co-workers on:
https://www.psi.ch/de/zrw
More information about Prof. Schibli and co-workers on:
https://www.psi.ch/de/zrw

Philipp Spycher, PhD is a post-doc in the laboratory of Prof. Roger Schibli at the Center for Radiopharmaceutical Science (CRS) at PSI Villigen, Switzerland since mid-2014. He attended his undergraduate studies at the University of Basel and ETH Zurich in Nanosciences and Biodmedical Engineering. He finished his PhD thesis in the laboratory of Prof. Viola Vogel (ETH Zurich) on engineering supported lipid membranes for cell biological applications; this included surface-micropatterning and enzymatic surface functionalization with cell-adhesive proteins using transglutaminase factorXIII. During his PhD he worked for several months in the laboratory of Prof. Erik Reimhult (BOKU, Vienna/Austria) learning about surface analysis tools and recombinant protein expression. He is interested in the development and application of new enzymatic bioconjugation technologies at the interface of biology, material science and medicine.
 Discover Our New Catalogue Edition and Dive into the World of Transglutaminases!
Discover Our New Catalogue Edition and Dive into the World of Transglutaminases!  Successful ISO9001:2015 recertification
Successful ISO9001:2015 recertification  Dr. Falk Pharma and Zedira announce successful completion of the phase 2a proof-of-concept study of ZED1227 for the treatment of Celiac Disease
Dr. Falk Pharma and Zedira announce successful completion of the phase 2a proof-of-concept study of ZED1227 for the treatment of Celiac Disease  Dr. Falk Pharma und Zedira verkünden den erfolgreichen Abschluss der Phase 2a-Studie mit ZED1227 zur Behandlung von Zöliakie
Dr. Falk Pharma und Zedira verkünden den erfolgreichen Abschluss der Phase 2a-Studie mit ZED1227 zur Behandlung von Zöliakie  Reversibly acting transglutaminase 2 inhibitors: drug candidates for the treatment of fibrosis
Reversibly acting transglutaminase 2 inhibitors: drug candidates for the treatment of fibrosis  Transglutaminase antibodies and neurological manifestations of gluten sensitivity
Transglutaminase antibodies and neurological manifestations of gluten sensitivity  Design of Oral FXIIIa Blockers as Safer Anticoagulants Mission Impossible?
Design of Oral FXIIIa Blockers as Safer Anticoagulants Mission Impossible?  Microbial transglutaminase (MTG) enables efficient and site-specific conjugation to native antibodies without the need of antibody engineering
Microbial transglutaminase (MTG) enables efficient and site-specific conjugation to native antibodies without the need of antibody engineering  Tridegin as FXIIIa inhibitor
Tridegin as FXIIIa inhibitor  Microbial transglutaminase: from discovery to market
Microbial transglutaminase: from discovery to market  Tissue transglutaminase inhibitors
Tissue transglutaminase inhibitors  Tissue transglutaminase in Alzheimers Disease
Tissue transglutaminase in Alzheimers Disease  Factor XIIIa: novel target for anticoagulation?
Factor XIIIa: novel target for anticoagulation?  Microbial transglutaminase for site-specific protein conjugation
Microbial transglutaminase for site-specific protein conjugation