Due to our proprietary laser printer-based on-chip synthesis, PEPperPRINT is able to routinely translate antigens into peptide microarrays with a maximum peptide-peptide overlap, e.g., 14 amino acids with 15mer peptides (equals a shift of 1), to provide the maximum amount of epitope coverage and prediction at reasonable costs.
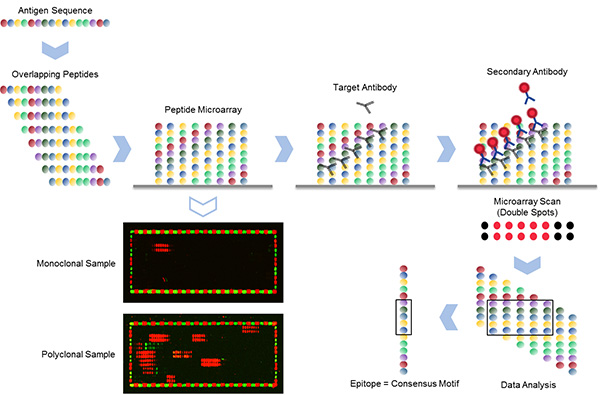
High-resolution linear epitope mapping is based on an antigen translated into 15 aa overlapping peptides with maximum peptide-peptide overlap of 14 aa (shift 1). The peptides are directly synthesized on the glass slides. The array is subsequently stained with the target antibody that binds to its specific epitope and a secondary antibody that has a fluorescence label. An image of the array is taken with a microarray scanner. Since PEPperPRINT prints spot duplicates, each signal appears in a double row. After data acquisition, the PepSlide® Analyzer software immediately calculates the common motif in the peptide hits, which is the epitope of the target antibody.
On the left, two examples of such an epitope mapping are shown, a monoclonal sample binding to a single epitope and a polyclonal sample binding to multiple epitopes. Each array is surrounded by HA (red) and FLAG (green) control peptides.
 Successful ISO9001:2015 recertification
Successful ISO9001:2015 recertification  Besuch des Bundesministers für Wirtschaft und Klimaschutz Dr. Robert Habeck bei der Zedira
Besuch des Bundesministers für Wirtschaft und Klimaschutz Dr. Robert Habeck bei der Zedira  Discover Our New Catalogue Edition and Dive into the World of Transglutaminases!
Discover Our New Catalogue Edition and Dive into the World of Transglutaminases!  Successful ISO9001:2015 recertification
Successful ISO9001:2015 recertification  Dr. Falk Pharma and Zedira announce successful completion of the phase 2a proof-of-concept study of ZED1227 for the treatment of Celiac Disease
Dr. Falk Pharma and Zedira announce successful completion of the phase 2a proof-of-concept study of ZED1227 for the treatment of Celiac Disease  Dr. Falk Pharma und Zedira verkünden den erfolgreichen Abschluss der Phase 2a-Studie mit ZED1227 zur Behandlung von Zöliakie
Dr. Falk Pharma und Zedira verkünden den erfolgreichen Abschluss der Phase 2a-Studie mit ZED1227 zur Behandlung von Zöliakie  Reversibly acting transglutaminase 2 inhibitors: drug candidates for the treatment of fibrosis
Reversibly acting transglutaminase 2 inhibitors: drug candidates for the treatment of fibrosis  Transcriptomic analysis of the efficacy of TG2-inhibitor trials and human intestinal organoids modelling Celiac disease pathogenesis
Transcriptomic analysis of the efficacy of TG2-inhibitor trials and human intestinal organoids modelling Celiac disease pathogenesis  Transglutaminase antibodies and neurological manifestations of gluten sensitivity
Transglutaminase antibodies and neurological manifestations of gluten sensitivity  Design of Oral FXIIIa Blockers as Safer Anticoagulants Mission Impossible?
Design of Oral FXIIIa Blockers as Safer Anticoagulants Mission Impossible?  Microbial transglutaminase (MTG) enables efficient and site-specific conjugation to native antibodies without the need of antibody engineering
Microbial transglutaminase (MTG) enables efficient and site-specific conjugation to native antibodies without the need of antibody engineering  Tridegin as FXIIIa inhibitor
Tridegin as FXIIIa inhibitor  Microbial transglutaminase: from discovery to market
Microbial transglutaminase: from discovery to market  Tissue transglutaminase inhibitors
Tissue transglutaminase inhibitors  Tissue transglutaminase in Alzheimers Disease
Tissue transglutaminase in Alzheimers Disease  Factor XIIIa: novel target for anticoagulation?
Factor XIIIa: novel target for anticoagulation?  Microbial transglutaminase for site-specific protein conjugation
Microbial transglutaminase for site-specific protein conjugation